3-D Molecules Move into the Fast Lane
Seeing compounds in three dimensions clears a path for making medicines faster, better.
Understanding how medicines behave around human cells was once a matter of educated guesswork, in large part because how they act at the molecular level isn’t visible. But today, structural biology and computational chemistry – or the use of computer modeling and molecular dynamics simulation – take medicine design into the next dimension. Well, to be precise, the third dimension.
With the jump into 3-D, chemists and biologists can precisely visualize the way molecules bind to the proteins that signal reactions in the body, such as the receptor protein for dopamine, a neurotransmitter that influences movement and regulates emotions. An imbalance of dopamine can be linked to neurological disorders like Parkinson’s disease and schizophrenia.
Locks and Keys
In practical terms, 3-D technology allows medicinal chemists like Anabella Villalobos, an expert in neuroscience medicinal chemistry at Pfizer's Groton, Connecticut site, to better make a medicine that can bind to natural gaps in receptor sites of proteins, providing the most effective treatment possible. “In 3D, we can get a picture of not only the protein but also of how the potential medicines fit,” said Villalobos.
When the newly created chemical compound—in this case, a potential medicine—binds, it produces a change in the shape of the protein. In general, this binding has one of two effects. It can “unlock” the door to an activity that would be beneficial to the patient, for example, by enhancing the activity of dopamine receptors in patients with Parkinson’s disease. This kind of compound is called an agonist. Or it can bind in such a way that it “locks” the door to an activity that is hindering the patient, such as by turning off the dopamine receptor in patients with schizophrenia or bipolar disorder. These compounds are called antagonists.
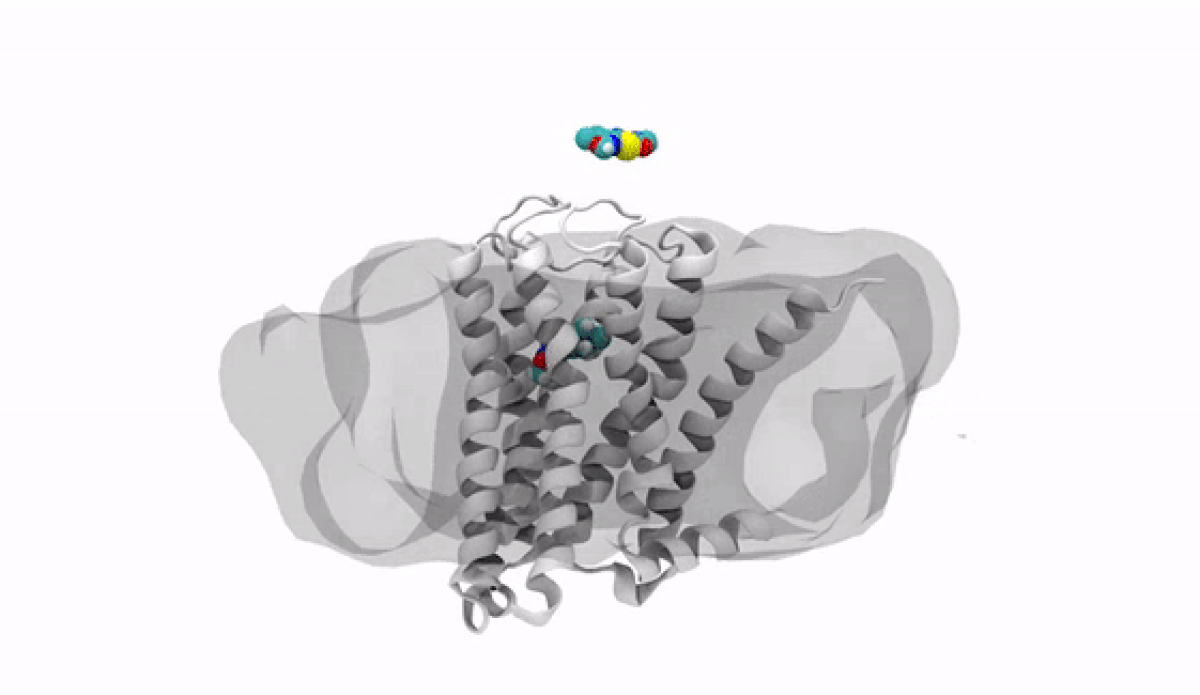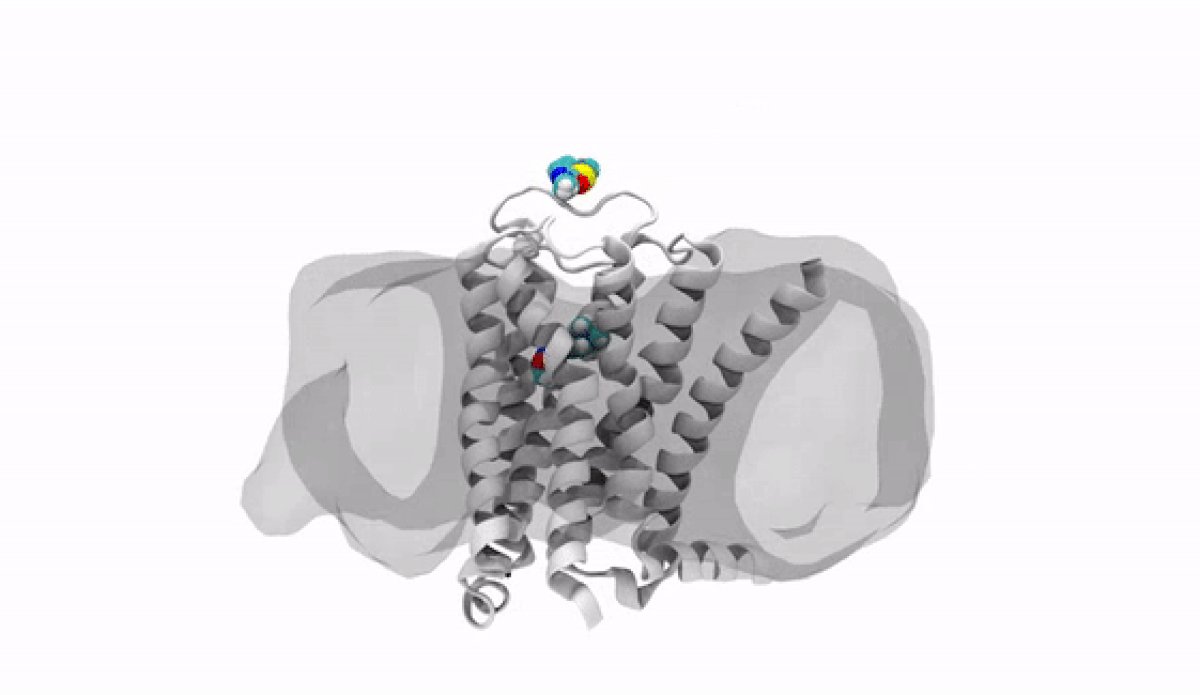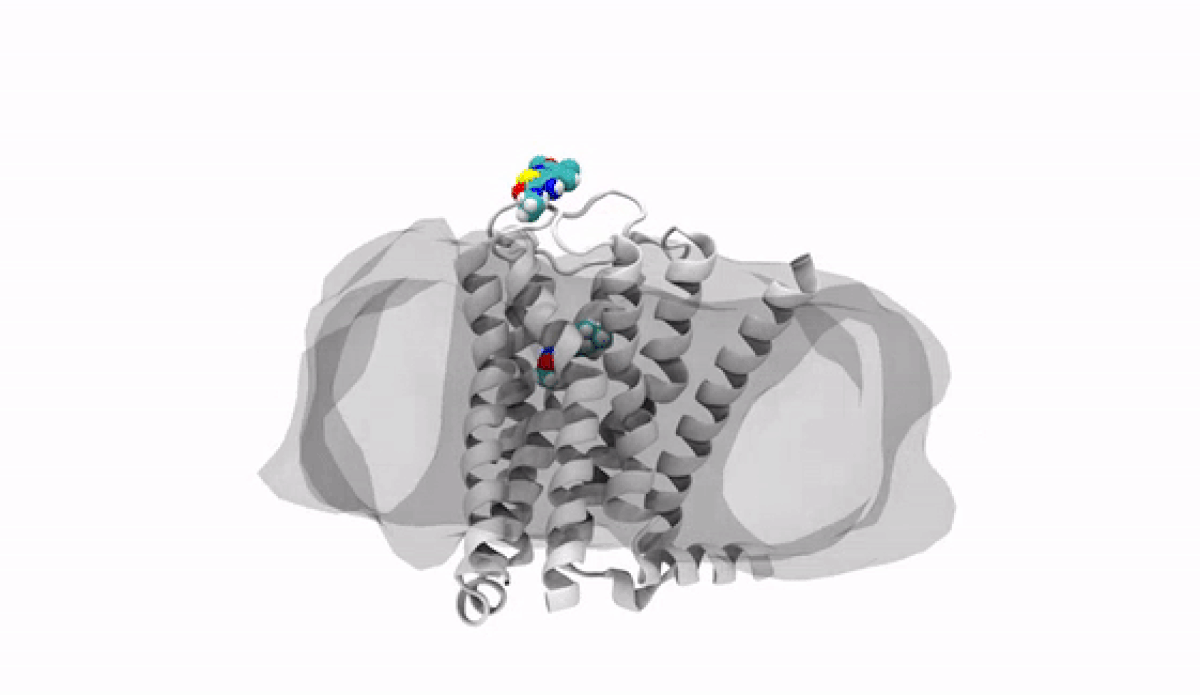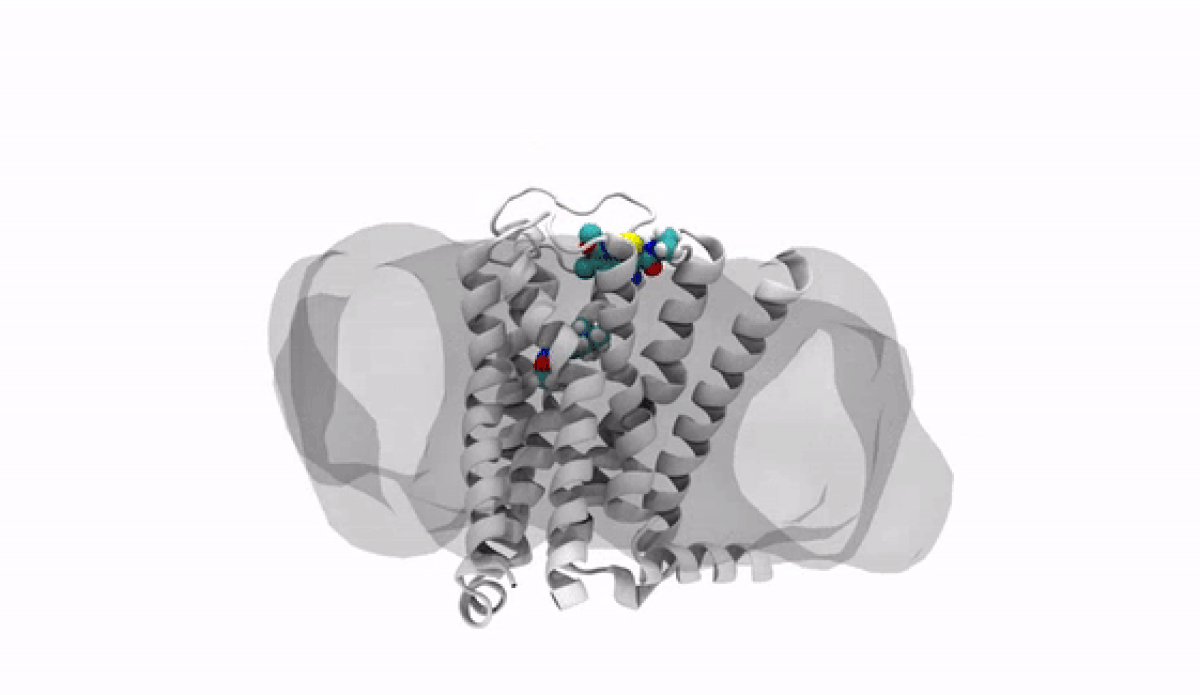
The Making of a 3-D Molecule
The road to 3-D molecular viewing starts with structural biologists who freeze and crystallize the protein to make its exact structure visible in a process called X-ray crystallography. Enter the computational chemists, such as Xinjun Hou, Director in Neuroscience and Pain Medicinal Chemistry at Pfizer's Kendall Square, Cambridge, Massachusetts site, who re-envisions them as a computerized molecule in three dimensions that the researchers can manipulate, twist, even turn on its head with a program called MoViT (Molecular Visualization Tool). To find the perfect fit for a particular molecule’s nooks and crannies, researchers start by poring over a database of millions of potential medicinal compounds, looking for good candidates. They then filter these down to a few thousand and follow up on a few hundred that look the most promising.
By tweaking a compound, perhaps adding an atom here or snipping one off there, Villalobos and the team can figure out the optimum complement for the receptor. The closer the fit, the more time the medicine will stay snuggled into the receptor. In practical terms, the team’s medicine design tweaks could potentially mean going from four doses a day down to a single dose by increasing the amount of time it takes for the body to break the medicine down.
It’s all because of a design methodology based on precise molecular structure, a frontier that 3-D modeling has made possible. “In the past, we didn’t have a picture of how the molecules bound to protein targets,” Villalobos said. “We’d keep trying various approaches then compare the results. That took a lot of time, and we had to make a lot of compounds.”
This 3-D method could mean discovering a medicine in less time, which translates to getting better drugs to the patients who need them, faster.